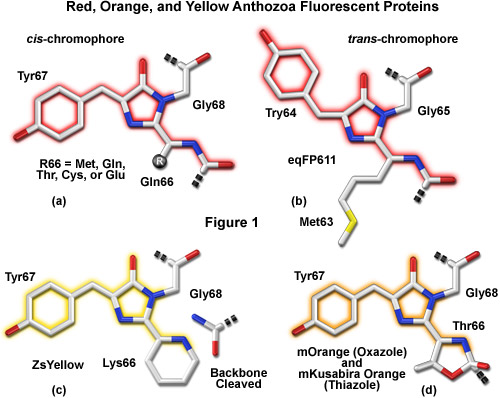
A variety of novel fluorescent proteins in virtually every color class have recently been cloned from Anthozoa and copepods, as well as a unique discovery of a family of GFPs in amphioxus. Researchers have extensively engineered these new probes to improve their utility for live-cell imaging. Essential to the understanding of spectral diversity in the wide range of fluorescent proteins discovered thus far are structural investigations of the stereochemical nature of the chromophore and the effects of its surrounding environment on fluorescent properties. Unlike the Aequorea GFP derivatives, there appears to be a high degree of variation in the chromophores of red-shifted fluorescent proteins (Figure 1). Thus, even through the DsRed chromophore configuration, termed planar cis (Figure 1(a)), appears to be the predominant structure in most proteins that emit in the orange and red regions, there are at least two additional motifs, planar trans and non-planar trans, which have been elucidated through x-ray diffraction studies.

The tutorial initializes with an image of the pre-maturation eqFP611 chromophore tripeptide amino acid sequence (Met63-Tyr64-Gly65) stretched into a linear configuration so that the methionine residue is positioned at the extreme left end of the window. Oxygen atoms are colored red, nitrogen atoms blue, carbon atoms white, the sulfur atom yellow, and the black dashes at the peptide termini indicate continuation of the backbone beyond the portion illustrated. In order to operate the tutorial, use the Chromophore Maturation Control slider to transition through the intramolecular rearrangement of the tripeptide sequence that occurs during chromophore maturation. The first step is a series of torsional adjustments that relocate the carboxyl carbon of Met63 in close proximity to the amino nitrogen of Gly65. Nucleophilic attack on this carbon atom by the amide nitrogen of glycine, followed by dehydration, results in formation of an imidazolin-5-one heterocyclic ring system. Green fluorescence emission from the immature chromophore (indicated by a green glow surrounding the affected structural elements) occurs when oxidation of the tyrosine alpha-beta carbon bond by molecular oxygen extends conjugation of the imidazoline ring system to include the tyrosine phenyl ring and its para-oxygen substituent. Proceeding with the maturation sequence to form the red chromophore (indicated by a red glow surrounding the affected structural elements), a second oxidation step involving the alpha-carbon and amide nitrogen of Met63 further increases the extended π-bonding electron resonance system to include the carboxyl group of Phe62. Adding this acylimine moiety to the chromophore results in a greater degree of electron delocalization during excitation, which partially accounts for the dramatic red shift of emission wavelengths observed in eqFP611 as well as DsRed and related fluorescent proteins.
A planar trans motif is found in the chromophore of the red fluorescent protein eqFP611, isolated from the sea anemone Entacmaea quadricolor (Figure 1(b)), which displays one of the largest Stokes shifts and red-shifted emission wavelength profiles of any naturally occurring Anthozoan fluorescent protein. In marked contrast, the non-planar trans conformation is characteristic of the non-fluorescent chromoprotein Rtms5 from Montipora efflorescens. In addition to the stereochemical variations, several of the proteins isolated from Anthozoa have markedly different chromophore structures from those of Aequorea GFP variants. The maturation of orange and red Anthozoa fluorescent proteins is believed to follow the same initial pathway as Aequorea GFP, but development continues with a second oxidation step that generates an acylimine moiety integrated into the peptide backbone between the amino acid residue preceding the chromophore and the first residue of the chromophore (usually methionine, glutamine, cysteine, or glutamic acid in Anthozoa fluorescent proteins). In several cases, the first residue of the chromophore also undergoes a cyclization reaction to form a third ring system (Figures 1(c) and 1(d)), which further influences the emission spectrum.
As further studies into the complex characteristics of fluorescent protein chromophores yield clues about the structure-function relationship with the polypeptide backbone, the task of genetically engineering more finely-tuned color variants and broadening the spectral range of useful proteins will undoubtedly become easier. Given that many of the amino acid triplets so far uncovered in chromophores from Anthozoa can give rise to huge variations in emission color (for example: MYG, 177 nanometers; QYG, 137 nanometers; TYG, 91 nanometers; CYG, 80 nanometers), it appears that there should be sufficient room in the fluorescent protein sequence space for additional mutations that will optimize color and many, if not all, of the other properties in fluorescent proteins derived from these organisms.
Crystallographic studies indicate that eqFP611 forms a tetramer exhibiting 222 symmetry, which is similar to that observed for the closely related DsRed fluorescent protein and also the non-fluorescent chromoprotein Rtms5. However, within the beta-barrel fold, the eqFP611 chromophore adopts a unique coplanar configuration in which the Try64 para-hydroxybenzylidene moiety is positioned trans rather than cis (as in DsRed) to the imidazoline ring. In addition, the bi-cyclic ring system and its substituents, which probably have a only limited mobility inside the protein, participate in numerous hydrogen bonds (at least nine) and a variety of non-polar van der Waals interactions with closely neighboring water molecules and amino acid residues. The extended hydrophobic methionine side chain fills a deep pocket that is also present in DsRed and Rtms5. Ultimately, the combined interactions between the chromophore and neighboring aliphatic and aromatic amino acid side chains (and water molecules) may be helpful in elucidating the mechanism behind the unusual fluorescent properties of the eqFP611 protein.
As mentioned above, the Anthozoan fluorescent protein eqFP611 exhibits bright far-red fluorescence with an excitation peak at 559 nanometers and a maximum emission wavelength of 611 nanometers, to produce one of the largest recorded Stokes shifts (approximately 52 nanometers) of any fluorescent protein. The quantum yield of fluorescence in eqFP611 is approximately 0.45, while the absorption spectrum has an extinction coefficient of 78,000 reciprocal moles per centimeter at 559 nanometers, values that combine to render the protein approximately as bright as EGFP. During the in vivo chromophore maturation process, which occurs in approximately 12 hours, the protein passes through a green intermediate state as described in the tutorial. After maturation, however, only a small fraction of this green species (less and 1 percent) can be detected. In contrast to other Anthozoan fluorescent proteins, eqFP611 has a reduced tendency to oligomerize at lower concentrations as evidenced through electrophoresis and single-molecule experiments, although at high concentrations, the protein does form tetramers. Site-directed mutagenesis efforts have yielded functional dimeric variants of eqFP611, and continued efforts led to a monomeric fluorescent protein (mRuby) from this species.
Contributing Authors
Tony B. Gines, Kevin A. John, Tadja Dragoo, and Michael W. Davidson - National High Magnetic Field Laboratory, 1800 East Paul Dirac Dr., The Florida State University, Tallahassee, Florida, 32310.




